CBTP in the news:
AAAS/CASE Workshop in DC features two CBTP fellows
Current Trainees

Danielle Cervasio
Program/Dates Supported by NIGMS: Chemistry/ July 2018-present
Title: Translational applications of small-molecule, astrocyte-targeted probes
Jinwoo Kim

Program/Dates Supported: Chemistry/ September 2017-present
Title: Total synthesis of (-)-incarvillateine and development of tumor-targeted drug delivery system
Amino acid is attached to acid-liable cleavable linker-embedded anti-cancer drug conjugate as a tumor targeting module (TTM). Cancer cells are known to overexpress specific amino acid transporters because of their high demand on nutrients for their rapid growth. Specific amino acid can guide anti-cancer agent toward cancer cells because it can be recognized as its transporter substrate. Once this drug conjugate is internalized via transporter-mediated endocytosis, low pH in tumor cells enables linker to get dissociated and release anti-cancer drug out inside tumor cells. Besides this research on drug conjugate, total synthesis of (-)-incarvillateine will be undertaken to elucidate mechanism of action for anti-inflammatory agents.
___________________________________________________________________________________
Jonathan Merino

Program/ Dates Supported on the NIGMS Grant: Chemistry/June 2017-present
Title: Synthesis of Substituted Benzoxaboroles to Target Bacterial DNA Ligase LigA via Substrate Assisted Tethered Inhibition
Benzoxaboroles are a class of drug-like molecules whose privileged scaffold has been exploited due to their excellent sugar-binding properties as well as improved water solubility, in contrast to the related phenylboronic acids. Tavaborole is an oxaborole used clinically to treat onychomycosis. The mechanism of action of tavaborole involves the formation of a covalent intermediate with leucyl-tRNA synthetase (LeuRS), an enzyme involved in protein synthesis. X-ray crystal structures of tavaborole bound to LeuRS demonstrate that the acidic oxaborole moiety forms an adduct with the cis-diol of the adenosine ribose of tRNALeu bound to LeuRS, trapping the substrate on the enzyme. The Tonge group posits that the specificity of the oxaboroles can be tailored so that they form adducts with the substrate diols in the enzyme-substrate complexes of other drug targets. This Substrate-Assisted Tethered Inhibition (SATI) approach is currently being employed to develop inhibitors of the NAD-dependent DNA ligase LigA, a target for the development of novel antibacterial agents. LigA is a promising target because the enzyme relies on NAD+ as its substrate as opposed to ATP in the human homolog (Lig1). Reaction of NAD with LigA leads to a covalent enzyme-AMP intermediate, and the goal of this project is to synthesize oxaboroles that form an adduct with the AMP diol in the LigA-AMP complex. Thus my research is focused on the development of methods to synthesize functionalized oxaboroles and to identify compounds that bind uncompetitively to LigA through the SATI mechanism LigA is a target in both Gram-positive and Gram-negative bacteria, and the long term goal of the project is to develop novel antibacterial agents that are active against drug-resistant nosocomial microorganisms.
____________________________________________________________________________________
Forrest Bowling

Program/Dates Supported on the NIGMS Grant: Biochemistry and Structural Biology (BSB) July 2017-present
Title: Regulation and Activation of Human Phospholipase D1
Phospholipase D1 (PLD1) generates phosphatidic acid and choline through the cleavage of phosphatidylcholine. PLD1 is involved in trafficking, endocytosis, and regulation of cell metabolism. PLD1 has an elevated activity level in transformed tumor cells and has been identified as a therapeutic target for cancer. Potent and specific small molecule inhibitors have been developed and pharmacological inhibition is effective in preventing cancer cell invasion in cell culture and neoangiogenesis in mice. Biochemically, PLD1 has low basal activity and requires activation by the anionic lipid PI(4,5)P2 through an interaction within the catalytic region. The focus of my research will be to visualize the mechanism of activation and inhibition of human PLD1 by lipids and small molecule inhibitors. My preliminary data has identified two baculovirus constructs of PLD1 that when expressed in Sf9 cells are soluble and ideal for structural studies. Utilizing structural information and biochemical assays, the mechanism of activation of human PLD1 will be determined. In addition, this structural information will be used to aid design of new inhibitors with increased half-lives as tool compounds for dissecting the benefits PLD inhibition in preventing cancer metastasis.
_______________________________________________________________________________
Ilana Heckler
Program/Dates Supported on the NIGMS Grant: Chemistry/ 2016-2018
Title: Nitric Oxide Mediated Bacterial Biofilm Dispersal: Investigation of a Nitric Oxide Sensing Phosphodiesterase from Vibrio cholerae
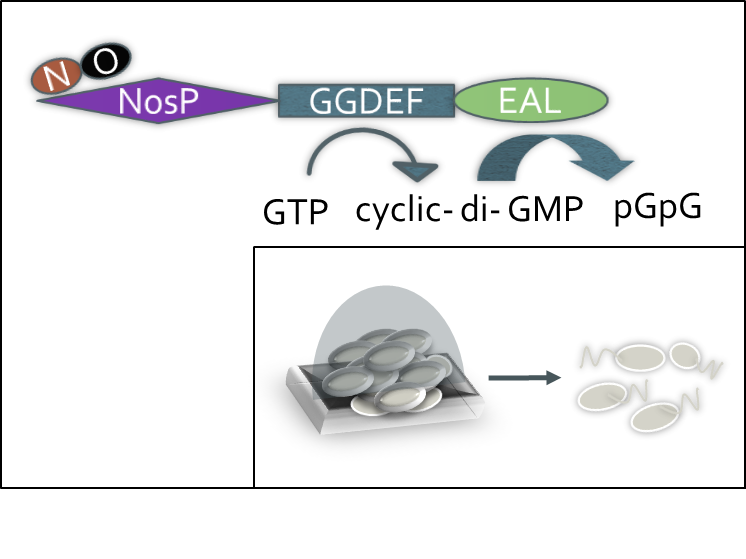
Bacterial biofilms are non-motile communities of bacteria. They are up to 1000-fold more resistant to antibiotics than free-swimming planktonic cells. Nitric oxide (NO) is small diatomic gas capable of diffusing through the protective matrix of biofilms, and has been shown to cause biofilm dispersal at nanomolar concentrations. The Boon lab has discovered a putative, novel NO sensing domain (NosP) that coordinates NO at a heme cofactor. This domain, like other sensor domains, is often encoded with catalytic domains. NosP is thought to interact with the catalytic domains responsible for the breakdown and synthesis of cyclic diguanylate monophosphate (c-di-GMP), the secondary messenger involved in bacterial motility, virulence and biofilm. The intracellular concentration of c-di-GMP is controlled by two enzymatic activities. Diguanaylate cyclase (DGC) activity is responsible for the synthesis of c-di-GMP through cyclization of two GTP molecules, and is catalyzed by a GGDEF motif. The breakdown of c-di-GMP into either pGpG or two molecules of GMP, by a phosphodiesterase (PDE), is catalyzed by an EAL or HD-GYP motif, respectively. The goal of this work is to elucidate the role that NO plays in controlling the intracellular concentration of c-di-GMP and therefore, motility, biofilm and virulence in Vibrio cholerae, the Gram-negative bacterium that is the causative agent of the intestinal infection cholera. V. cholerae encodes 22 phosphodiesterases, one of which is fused to a NosP domain. We propose that NO, upon binding to NosP, will regulate the transcription of genes involved in biofilm and virulence though activation of the hydrolytic activity of the PDE.
____________________________________________________________________________________

Stephen Collins
Program/Dates supported on the NIGMS Grant: Biochemistry and Structural Biology/2016-2017
Title: Inhibitor Development for Cancer-Associated Mutants of Human Epidermal Growth Factor Receptor 2 (HER2)
A subset of human epidermal growth factor receptor 2 (HER2) mutations uncovered by breast cancer genome sequencing and confirmed by the Miller lab cause deregulated kinase activity and signaling. These mutations likely represent driver mutations in breast cancer. By working with Jiaye Gou and Robert Rizzo a model of the active form of HER2 has been developed based on crystallographic information of epidermal growth factor receptor (EGFR) a HER2 family member with significant sequence homology. This had to be done because there is no structural information available for the active form of the kinase. Using this model and the zinc database we have been able to purchase a set of 149 compounds which represent our best chance at inhibiting HER2. By utilizing inhibitors as a chemical tools we aim to gain a more complete understanding of active HER2 which is poorly understood compared to EGFR.
____________________________________________________________________________________
Eric Roth

Program/Dates Supported on the NIGMS Grant: Molecular and Cellular Pharmacology/2016-2018
Title: Roles of Phospholipase D1 and D2 in PyMT driven Breast Cancer Metastasis
The goal of my research is to elucidate the effects of phospholipase D (PLD) in breast cancer metastasis and metabolism to utilize and develop compounds that target metastasis. PLD is expressed in two predominant isoforms, PLD1 and PLD2, both of which generate the lipid signaling molecule phosphatidic acid through the hydrolysis of phosphatidylcholine. Both PLD1 and PLD2 have been shown to promote several early metastatic steps including migration and invasion from the primary tumor, neoangiogenesis, and survival in circulation. Although highly related, their cellular roles are not always redundant or additive, but can also be contradictory. As such, it is necessary to elucidate their separate and combined effects. In order to investigate the effects of dual inhibition, I am using the PLD1/2 inhibitor FIPI and mice genetically ablated of PLD1/2 producing a double knockout (DKO). The C57/B6 mice develop tumors from the polyoma middle-T antigen (PyMT) under expression of the mouse mammary tumor virus (MMTV). These MMTV-PyMT mice spontaneously develop multifocal mammary adenocarcinomas that regularly metastasize to the lung and lymph nodes. Previous studies have shown that PLD1-/- mice exhibit reduced metastasis relative to wild-type mice. My preliminary data shows a further reduction in DKO mice that results in a dramatic suppression of lung metastasis. Furthermore, DKO mice have decreased tumor burden compared to both wild-type and PLD1-/- mice. In addition to suppressing metastasis, inhibition of PLD1 has also been shown to disrupt neoangiogenesis, autophagy, and lipid droplet formation. Collectively, this results in limited metabolic flexibility and energy reserves of tumor cells while also exposing them to hypoxic conditions. I expect these cells develop focused metabolic dependencies not present in normal cells, providing an opportunity for combined inhibition to produce synthetic lethality.
___________________________________________________________________________________
Lauren Prentis
Program/Dates Supported on the NIGMS Grant: Chemistry/ 2016-2018
Title: Computational De Novo design of small molecule inhibitors for viral fusion proteins in Ebola and HIV
The goal of my proposed research is to develop and apply improved atomic level computational modeling tools to clinically relevant drug targets. The research integrates chemistry, biology, and physics-based approaches and is directly relevant to the field of Chemical Biology. My initial efforts have focused on targeting the Ebola virus (EBOV), which infects its host through fusion with the host cell membrane after endocytosis and is fatal. The glycoprotein on the EBOV surface inserts into the host membrane and as the pH drops to an acidic environment, the glycoprotein subunit 2 (GP2) changes its conformation from a mostly disordered to a stable 6 helix bundle. This structural change is thought to overcome the energetic barrier associated with lipid mixing that allows membrane and ultimately viral entry. The aim of this work is to design a small molecule candidate to bind to an outer pocket on GP2 delaying the formation of the 6 helix bundle, allowing the EBOV to be lysed by the cell. The project involves design, optimization, and use of a genetic algorithm approach that employs de novo design principles to breed small molecules for compatibility with a protein binding site. Through the use of the genetic algorithm approach, the offspring molecules will be selected as parents for following generations with different selection methods that employ the use of different scoring functions to rank and select the ensemble. The goal is to create a novel population of molecules with enhanced binding potential compared to an initial ensemble. Importantly, the genetic algorithm approach can be used across various drug targets and although my initial efforts have focused on EBOV, I plan to apply the method to other drug discovery projects in the lab targeting HIV gp41 and fatty acid binding protein (FABP). We anticipate the use of a genetic algorithm approach in combination with our current virtual screening approach that employs docking, will be a more effective way to identify new drug leads for important drug targets.
____________________________________________________________________________________
Matthew Cifone

Program/Dates Supported on the NIGMS Grant: Chemistry/ 2016-2018
Title: Design and Synthesis of Bivalent Inhibitors Against Acetyl-CoA Carboxylase.
A major issue currently facing the pharmaceutical industry is the rise in bacterial strains that are resistant to commercially available antibiotics. There is thus an urgent need to identify new drug targets. My research project is focused on the enzyme complex acetyl-CoA carboxylase (ACC), which participates in the first committed step of fatty acid biosynthesis and is composed of two enzymes, biotin carboxylase (BC) and carboxyltransferase (CT), as well as biotin carboxyl carrier protein (BCCP). This complex is particularly attractive because ACC is encoded differently between prokaryotes and eukaryotes, allowing us to selectively inhibit bacterial ACC. Selective and potent inhibitors of both the BC and CT domains have already been developed including the pyridopyrimidines that target the BC domain and moiramide B which inhibits the CT domain. The goal of my project is to develop bivalent inhibitors of ACC by linking the BC and CT pharmacophores using a PEG linker. The bivalent inhibitors are excepted to have higher affinity for the enzyme than either pharmacophore alone, and are also anticipated to have long residence time on the enzyme. As such, the bivalent inhibitors will be valuable tool compounds for exploring the relationship between time-dependent enzyme inhibition and prolonged drug activity at low concentration. In addition, the bivalent compounds are also expected to have improved resistance profile than either ACC inhibitor alone. Finally, since no structure exists for the complete bacterial ACC complex, we hope that the bivalent inhibitors will stabilize the complex sufficiently to facilitate protein crystallography. Because there is no structure of the complete complex, and initial aim of my project is to experiment with linkers of different sizes to determine the optimum separation of the two pharmacophores to bridge the BC and CT domains.
___________________________________________________________________________________
James Iuliano
Program/Dates supported on the NIGMS Grant: Chemistry/2015-2017
Title: Ultrafast Kinetics and Structural Dynamics of Light, Oxygen, Voltage Domain Proteins.
Activated proteins provide a means of high spatial and temporal control needed for the emerging field of optogenetics, in which light is used to control biological function. The most common application of optogenetics is for activating and deactivating neurons but has recently been extended to control of enzyme activity and cell signaling. Protein fusions containing light, oxygen, voltage (LOV) domains have been used as optogenetic tools, although much remains to be resolved on the mechanism of signal transduction from the flavin mononucleotide chromophore of the LOV domain to the effector domain, which can bind ligands or have enzymatic activity. The LOV proteins YtvA from Bacillus subtilis and EL222 from Erythrobacter litoralis are directly fused to effector domains through an alpha helix linker, providing model systems for studying initial light activation from the ultrafast (femtosecond) to millisecond timescales. We employ the use of ultrafast infrared spectroscopy and X-ray crystallography to study chromophore-protein and inter-domain interactions that lead to active states in these proteins.
____________________________________________________________________________________

Amber Bonds
Program/Dates supported: Chemistry/ 2014 - 2016
Title: Understanding the role of post-translational modifications on cholesterol utilization in Mycobacterium tuberculosis
Tuberculosis, the disease caused by Mycobacterium tuberculosis, is the second leading cause of death worldwide. Recent years have witnessed increased incidences of drug resistant strains; demonstrating the imperative need for new antibiotics. M.tbcholesterol utilization has great potential as a new therapeutic target. During the course of infection, M.tb resides in the lipid-rich environment of foamy macrophages which contain cholesterol and cholesterol esters. To adapt to this lipid-rich environment, M.tbupregulates genes involved in lipid metabolism. The Sampson laboratory has elucidated the mechanisms of cholesterol degradation and extensively characterized a class of cholesterol degradative enzymes known as acyl-CoA dehydrogenases (ACADs). Aside from transcriptional regulation, it is suggested that M.tb can regulate metabolic flux through post translational modifications (PTMs). PTMs have been shown to negatively regulate the activity of various enzymes. A newly identified PTM, succinylation, occurs extensively in M.tb and 19 of the cholesterol metabolizing enzymes have been identified as targets of succinylation. This work investigates the effects of succinylation on cholesterol utilization in M.tb and demonstrates the succinylase activity of a previously annotated acetyl-transferase, Nat. This data suggests that succinylation plays a significant role in the regulation of cholesterol utilization by M.tb during the course of infection.
____________________________________________________________________________________
 Alyssa Preston
Alyssa Preston
Program/Dates supported on the NIGMS Grant: Chemistry/ 2014-2016
Title: Tracing the brain’s neural circuitry with small molecule probes.
The brain functions by using neurons that link together through synaptic connections
to form neural circuits. Discovering the structure of these circuits is extremely
challenging, partly because of the limited number of techniques available for visualizing
their anatomy. The most powerful existing strategies employ viruses and lectins that
can move across the synapse, allowing one to visualize neural circuits if the virus
or lectin is tagged with a fluorophore. However, these viruses and lectins are only
applicable to limited species, sluggish in their labelling of neural circuits, and
not easily modified to improve their functions. My research focuses on the design
and synthesis of small-molecule probes for use as trans-synaptic tracers of neural
circuitry. In order to design these molecules, I take advantage of the feature of
trans-synaptic lectins that enables them to cross the synapse: their affinity for
sugars at the cell surface. I hypothesize that small molecules with sugar binding
properties similar to trans-synaptic lectins will be able to cross the synapse. I
have recently completed the synthesis of a model fluorescent molecule with high sugar
affinity, and I am currently exploring its ability to trace neural circuits in the
olfactory system of larval zebrafish. 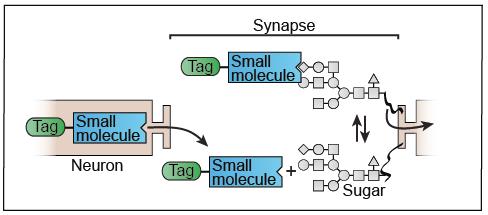
____________________________________________________________________________________
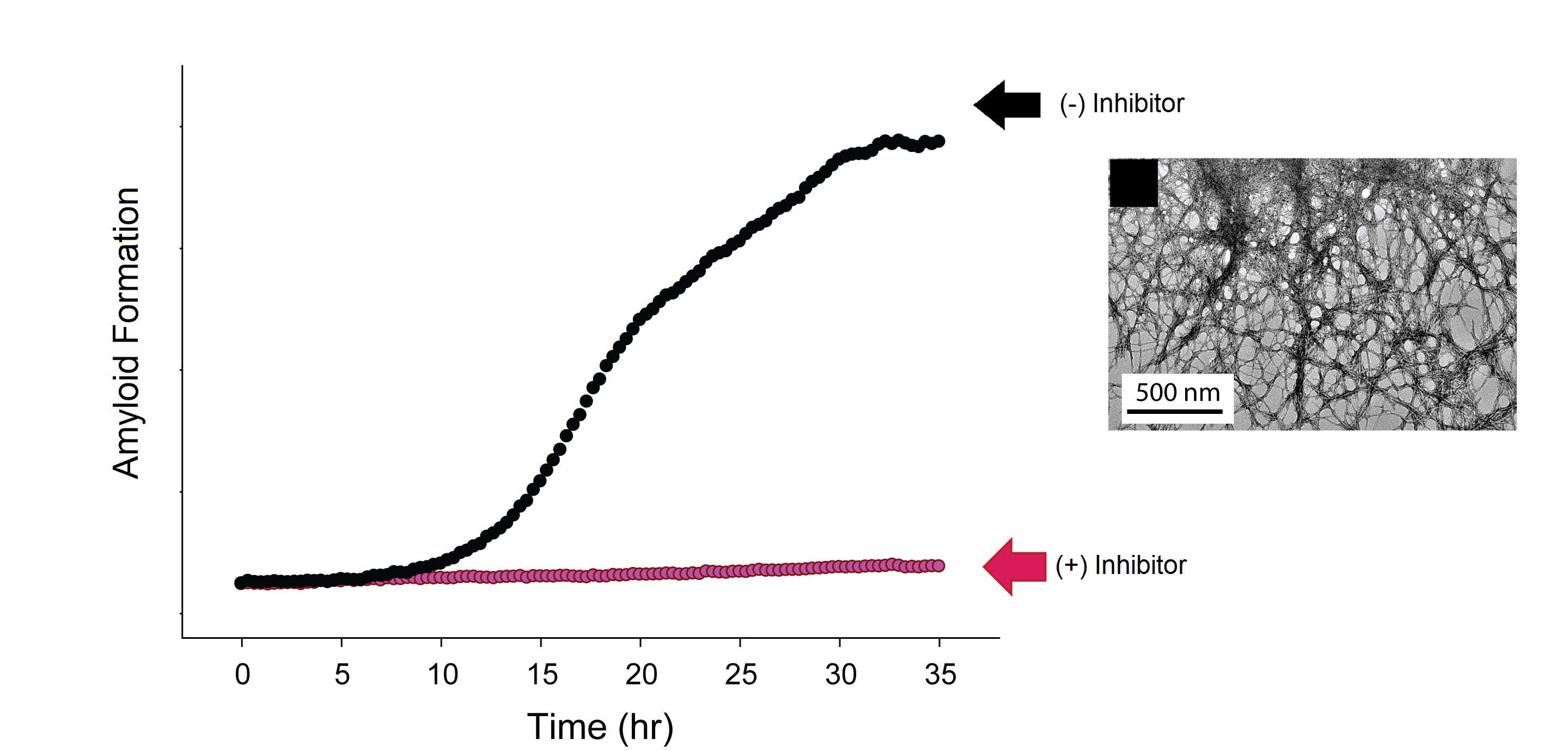
 Zachary Ridgway
Zachary Ridgway
Program/Dates supported on the NIGMS Grant: Chemistry/ 2014-2016
Title: Design and analysis of soluble IAPP analogs that act as amyloid inhibitors
Type II diabetes (T2D) is a disease that affects 6.0% of the world’s adult population. With no cure, there is a need to expand current methods of treatment. Upon the onset of T2D, a peptide hormone called IAPP (Islet amyloid polypeptide) misfolds to form insoluble fibrils. The aggregation of IAPP is associated with a decrease in beta cell mass and insulin production, contributing greatly to the progression of the disease. Soluble analogs of IAPP have been explored as a platform for therapeutic applications. By making key substitutions to the sequence of human IAPP, a class of analogs is created that act as inhibitors of IAPP aggregation. My research focuses on understanding the mechanisms by which these analogs inhibit cytotoxicity, through experimental and computational approaches. This will create a better understanding of the toxic intermediate species formed by IAPP, as well as determine criteria for designing more effective inhibitors.